The producers/consumers/reducers model
Who we are
- Gervasio Perez, MSc (PHD student)
- Sergio Yovine, Professor (advisor)
Our vision
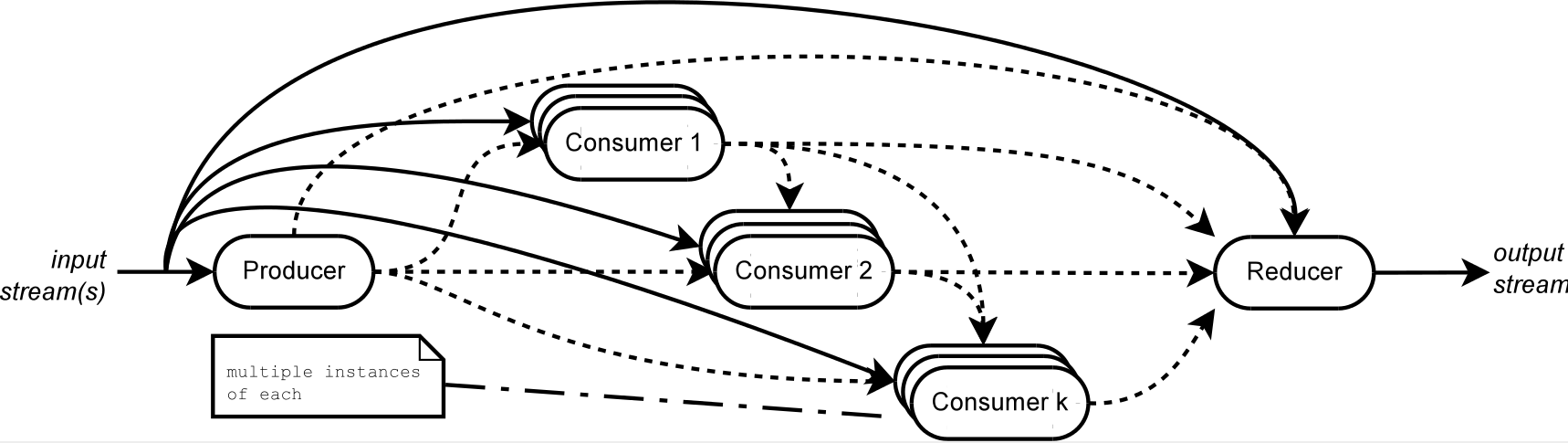
We propose the Producers/Consumers/Reducers model (PCR for short) as a declarative way of describing concurrent computation patterns and translating them into different implementations.
Our aim is to develop a flexible specification language along with a translation scheme allowing for multi-target parallel and distributed code generation.
The model
Atoms. In our model we identify as basic components a producer of data, consumers (or filters) of produced data, and a reducer combining one or more output streams back into a single result. All these components are connected by data streams. Business logic is written in the target language and bound to the corresponding PCR component.
Consumers and the reducer start working as soon as there are items to consume from the producer; there can be concurrent instances of each consumer.
The model supports reuse by allowing nesting of previously defined PCRs as consumer components of a newly defined one.
Example: Fibonacci Primes
Below we give a PCR description of the problem of finding Fibonacci numbers which are also prime.
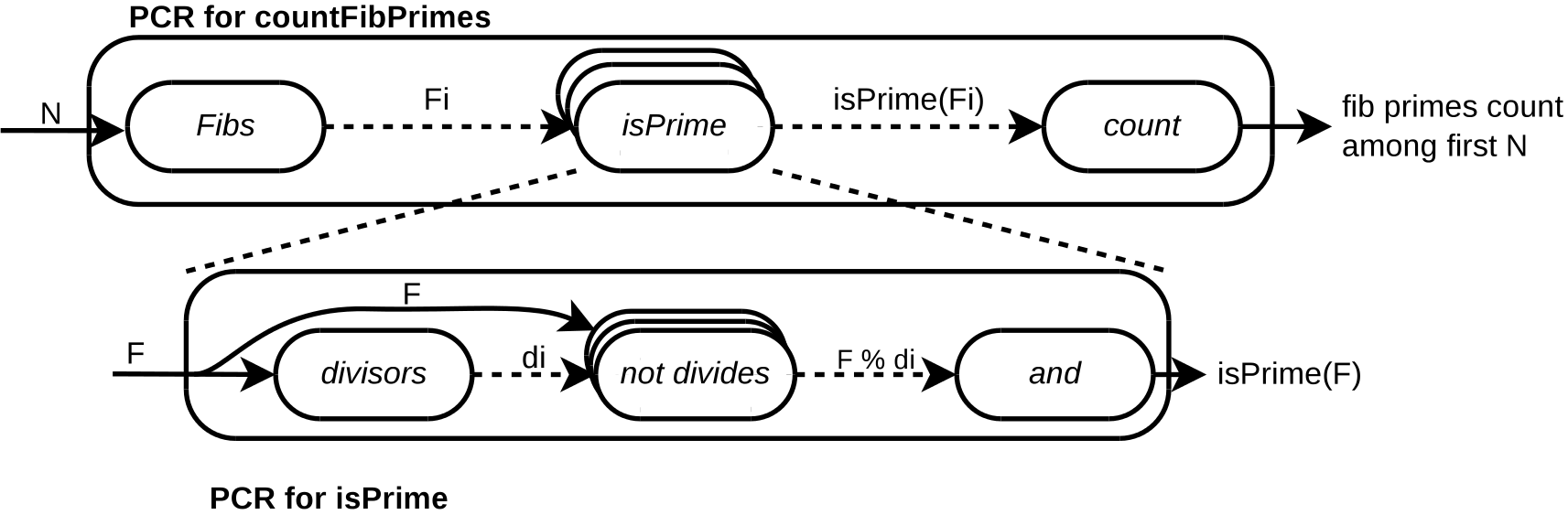
Intuitively, the Fibs producer reads N from the outer context, and generates the first N Fibonacci numbers. For each generated number, the isPrime consumer checks its primeness, producing a boolean value. Finally, the count reducer calculates the number of prime numbers found and outputs the result.
As shown, isPrime itself can be written as a PCR, allowing for reuse of its specification in multiple contexts.
A language for PCRs
The textual representation of the previous PCRs could be written as follows:
PCR isPrime(N):
par
d = produce divisors(N)
forall d:
y = consume not_divides(d,N)
p = reduce and true y
return p
PCR countFibPrimes(N):
par
f = produce Fibs(N)
forall f:
p = consume isPrime(f)
sum = reduce count 0 p
return sum
This language for describing PCRs and its formal model are based on the FXML coordination language.
Writing actual code: PCRs in C++
How would this look embedded in an actual programming language?
To answer this, we wrote a C++ template metaprogramming library to enable writing of PCRs:
typedef pcr< pcr_in <num_t>, pcr_produce <all_divisors, 1>, pcr_consume <not_divides, 1, 2>, pcr_reduce <And, 1> > par_is_prime; typedef pcr< pcr_in < int >, pcr_produce< Fibs, 1>, pcr_consume< par_is_prime, 1>, pcr_reduce < sum_primes, 1> > countFibPrimes;
This snippet shows the definition of PCRs as C++ types, abstracting out the actual implementation. Numbers indicate the source each producer/consumer/reducer parameter as a relative position in backwards steps.
A translation target: Intel Concurrent Collections
Having a way to express PCRs in C++ brings us to the task of providing translation targets. We chose C++ Intel Concurrent Collections (CnC) as a target execution model.
The CnC model consists in an execution graph connecting execution steps connected by item collections. Execution is controlled by tags used for signalling when new items are available for processing. The following figure shows an example calculating the primeness of each of the first N Fibonacci numbers.
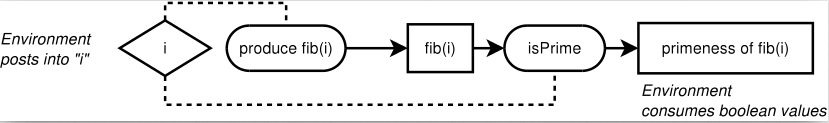
The Intel CnC implementation in C++ is based on the Intel TBB library. We wrote a CnC code generator for PCRs using template metaprogramming techniques.
More info about the CnC model in the Rice university project page.
Resources
- Link to the GIT repository with the current implementation.
- Unpublished whitepaper explaining the PCR model and technical details of code generation and mapping into the target (CnC) model.
- FXML info: A formal approach to derivation of concurrent implementations in software product lines

